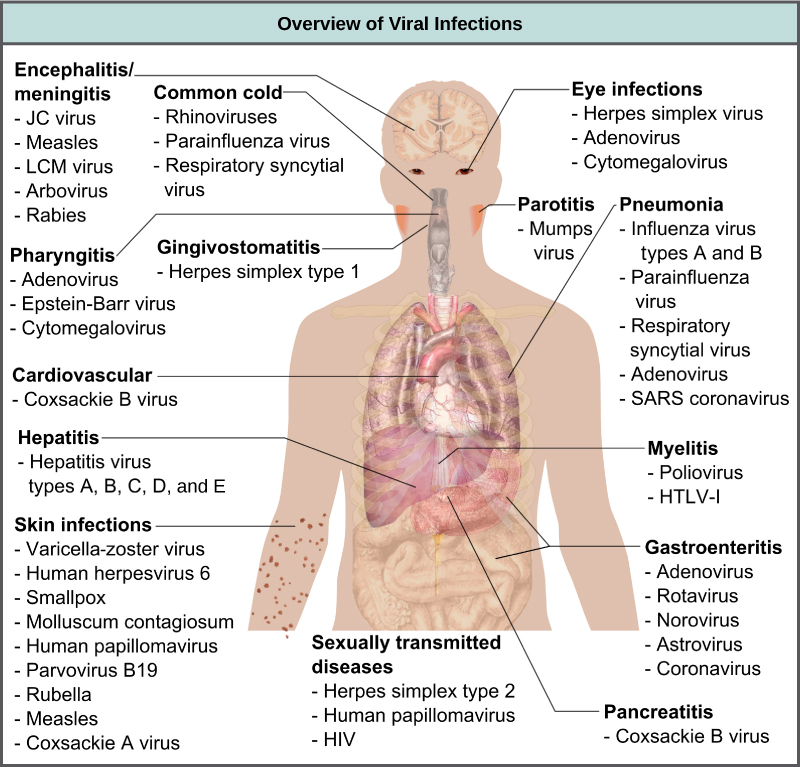
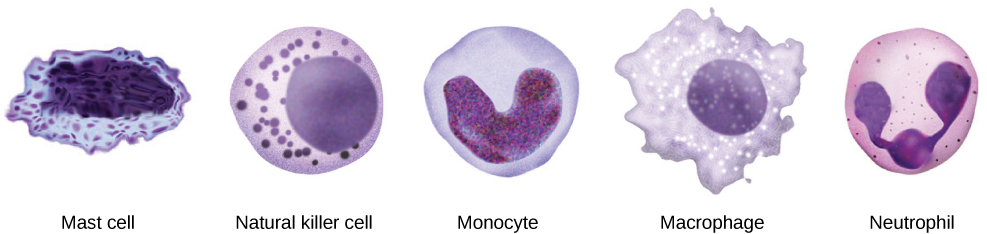
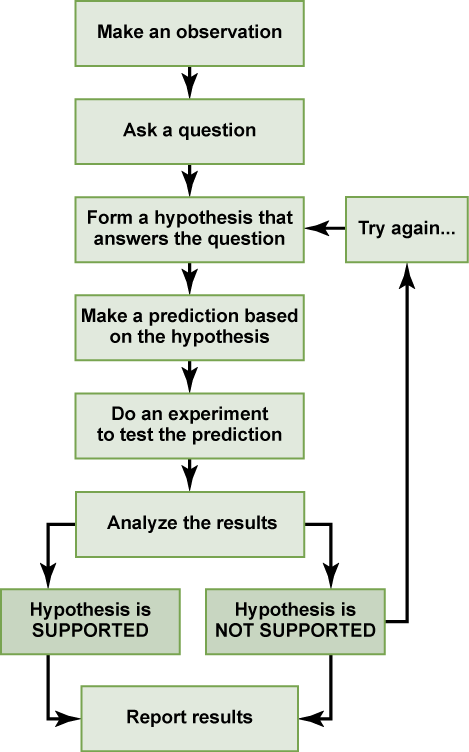
By the end of this section, you will be able to:
- Describe the ways in which carbon is critical to life
- Explain the impact of slight changes in amino acids on organisms
- Describe the four major types of biological molecules
- Understand the functions of the four major types of molecules
Watch a video about proteins and protein enzymes.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://opentextbc.ca/biology/?p=4368#h5p-14
The large molecules necessary for life that are built from smaller organic molecules are called biological macromolecules. There are four major classes of biological macromolecules (carbohydrates, lipids, proteins, and nucleic acids), and each is an important component of the cell and performs a wide array of functions. Combined, these molecules make up the majority of a cell’s mass. Biological macromolecules are organic, meaning that they contain carbon. In addition, they may contain hydrogen, oxygen, nitrogen, phosphorus, sulfur, and additional minor elements.
Carbon
It is often said that life is “carbon-based.” This means that carbon atoms, bonded to other carbon atoms or other elements, form the fundamental components of many, if not most, of the molecules found uniquely in living things. Other elements play important roles in biological molecules, but carbon certainly qualifies as the “foundation” element for molecules in living things. It is the bonding properties of carbon atoms that are responsible for its important role.
Carbon Bonding
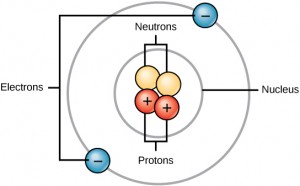
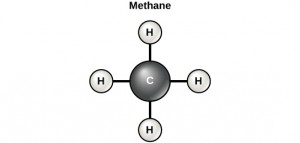
Carbon contains four electrons in its outer shell. Therefore, it can form four covalent bonds with other atoms or molecules. The simplest organic carbon molecule is methane (CH4), in which four hydrogen atoms bind to a carbon atom.
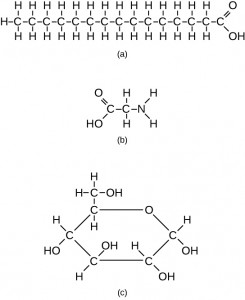
However, structures that are more complex are made using carbon. Any of the hydrogen atoms can be replaced with another carbon atom covalently bonded to the first carbon atom. In this way, long and branching chains of carbon compounds can be made (Figure 2.13 a). The carbon atoms may bond with atoms of other elements, such as nitrogen, oxygen, and phosphorus (Figure 2.13 b). The molecules may also form rings, which themselves can link with other rings (Figure 2.13 c). This diversity of molecular forms accounts for the diversity of functions of the biological macromolecules and is based to a large degree on the ability of carbon to form multiple bonds with itself and other atoms.
Carbohydrates
Carbohydrates are macromolecules with which most consumers are somewhat familiar. To lose weight, some individuals adhere to “low-carb” diets. Athletes, in contrast, often “carb-load” before important competitions to ensure that they have sufficient energy to compete at a high level. Carbohydrates are, in fact, an essential part of our diet; grains, fruits, and vegetables are all natural sources of carbohydrates. Carbohydrates provide energy to the body, particularly through glucose, a simple sugar. Carbohydrates also have other important functions in humans, animals, and plants.
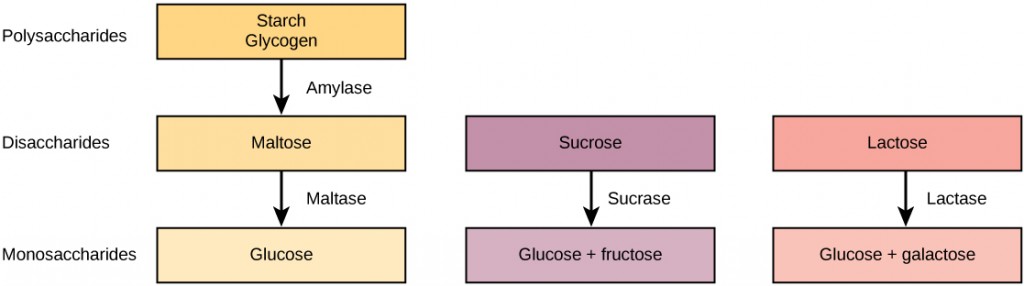
Carbohydrates can be represented by the formula (CH2O)n, where n is the number of carbon atoms in the molecule. In other words, the ratio of carbon to hydrogen to oxygen is 1:2:1 in carbohydrate molecules. Carbohydrates are classified into three subtypes: monosaccharides, disaccharides, and polysaccharides.
Monosaccharides (mono- = “one”; sacchar- = “sweet”) are simple sugars, the most common of which is glucose. In monosaccharides, the number of carbon atoms usually ranges from three to six. Most monosaccharide names end with the suffix -ose. Depending on the number of carbon atoms in the sugar, they may be known as trioses (three carbon atoms), pentoses (five carbon atoms), and hexoses (six carbon atoms).
Monosaccharides may exist as a linear chain or as ring-shaped molecules; in aqueous solutions, they are usually found in the ring form.
The chemical formula for glucose is C6H12O6. In most living species, glucose is an important source of energy. During cellular respiration, energy is released from glucose, and that energy is used to help make adenosine triphosphate (ATP). Plants synthesize glucose using carbon dioxide and water by the process of photosynthesis, and the glucose, in turn, is used for the energy requirements of the plant. The excess synthesized glucose is often stored as starch that is broken down by other organisms that feed on plants.
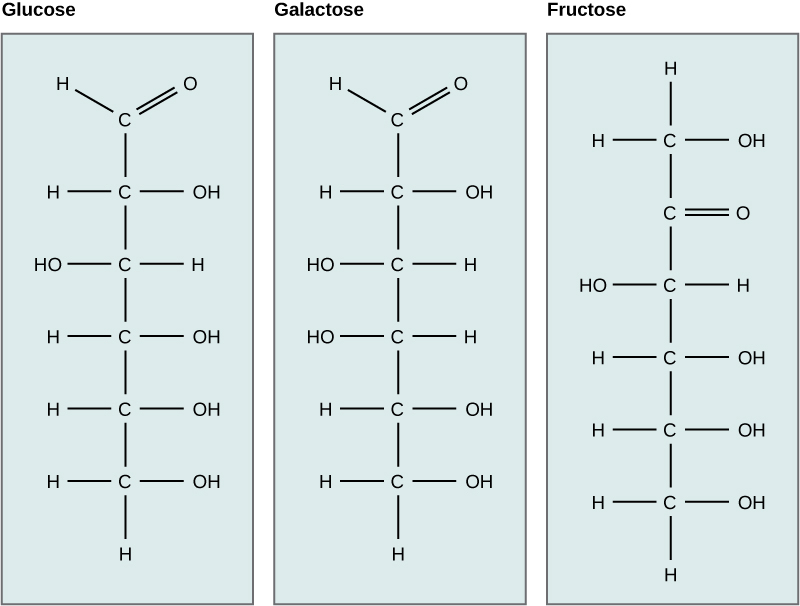
Galactose (part of lactose, or milk sugar) and fructose (found in fruit) are other common monosaccharides. Although glucose, galactose, and fructose all have the same chemical formula (C6H12O6), they differ structurally and chemically (and are known as isomers) because of differing arrangements of atoms in the carbon chain.
Disaccharides (di- = “two”) form when two monosaccharides undergo a dehydration reaction (a reaction in which the removal of a water molecule occurs). During this process, the hydroxyl group (–OH) of one monosaccharide combines with a hydrogen atom of another monosaccharide, releasing a molecule of water (H2O) and forming a covalent bond between atoms in the two sugar molecules.
Common disaccharides include lactose, maltose, and sucrose. Lactose is a disaccharide consisting of the monomers glucose and galactose. It is found naturally in milk. Maltose, or malt sugar, is a disaccharide formed from a dehydration reaction between two glucose molecules. The most common disaccharide is sucrose, or table sugar, which is composed of the monomers glucose and fructose.
A long chain of monosaccharides linked by covalent bonds is known as a polysaccharide (poly- = “many”). The chain may be branched or unbranched, and it may contain different types of monosaccharides. Polysaccharides may be very large molecules. Starch, glycogen, cellulose, and chitin are examples of polysaccharides.
Starch is the stored form of sugars in plants and is made up of amylose and amylopectin (both polymers of glucose). Plants are able to synthesize glucose, and the excess glucose is stored as starch in different plant parts, including roots and seeds. The starch that is consumed by animals is broken down into smaller molecules, such as glucose. The cells can then absorb the glucose.
Glycogen is the storage form of glucose in humans and other vertebrates, and is made up of monomers of glucose. Glycogen is the animal equivalent of starch and is a highly branched molecule usually stored in liver and muscle cells. Whenever glucose levels decrease, glycogen is broken down to release glucose.
Cellulose is one of the most abundant natural biopolymers. The cell walls of plants are mostly made of cellulose, which provides structural support to the cell. Wood and paper are mostly cellulosic in nature. Cellulose is made up of glucose monomers that are linked by bonds between particular carbon atoms in the glucose molecule.
Every other glucose monomer in cellulose is flipped over and packed tightly as extended long chains. This gives cellulose its rigidity and high tensile strength—which is so important to plant cells. Cellulose passing through our digestive system is called dietary fiber. While the glucose-glucose bonds in cellulose cannot be broken down by human digestive enzymes, herbivores such as cows, buffalos, and horses are able to digest grass that is rich in cellulose and use it as a food source. In these animals, certain species of bacteria reside in the rumen (part of the digestive system of herbivores) and secrete the enzyme cellulase. The appendix also contains bacteria that break down cellulose, giving it an important role in the digestive systems of ruminants. Cellulases can break down cellulose into glucose monomers that can be used as an energy source by the animal.
Carbohydrates serve other functions in different animals. Arthropods, such as insects, spiders, and crabs, have an outer skeleton, called the exoskeleton, which protects their internal body parts. This exoskeleton is made of the biological macromolecule chitin, which is a nitrogenous carbohydrate. It is made of repeating units of a modified sugar containing nitrogen.
Thus, through differences in molecular structure, carbohydrates are able to serve the very different functions of energy storage (starch and glycogen) and structural support and protection (cellulose and chitin).
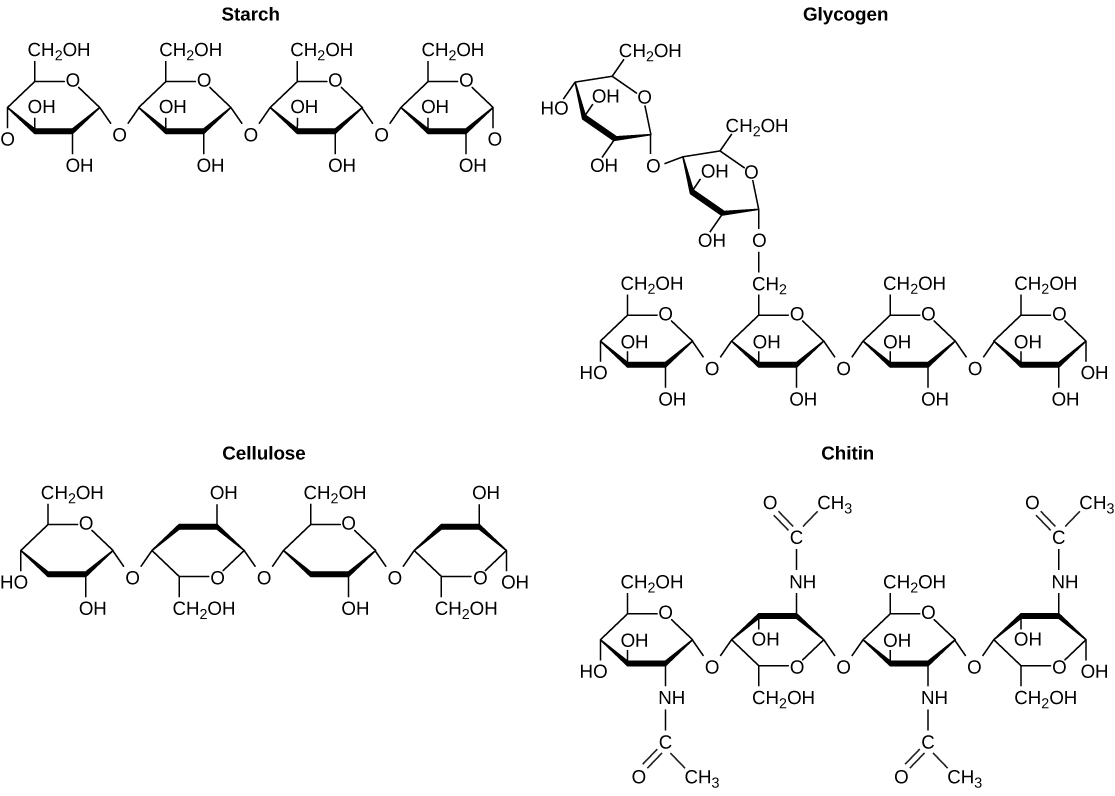
Figure 2.15 Although their structures and functions differ, all polysaccharide carbohydrates are made up of monosaccharides and have the chemical formula (CH2O)n.
Registered Dietitian: Obesity is a worldwide health concern, and many diseases, such as diabetes and heart disease, are becoming more prevalent because of obesity. This is one of the reasons why registered dietitians are increasingly sought after for advice. Registered dietitians help plan food and nutrition programs for individuals in various settings. They often work with patients in health-care facilities, designing nutrition plans to prevent and treat diseases. For example, dietitians may teach a patient with diabetes how to manage blood-sugar levels by eating the correct types and amounts of carbohydrates. Dietitians may also work in nursing homes, schools, and private practices.
To become a registered dietitian, one needs to earn at least a bachelor’s degree in dietetics, nutrition, food technology, or a related field. In addition, registered dietitians must complete a supervised internship program and pass a national exam. Those who pursue careers in dietetics take courses in nutrition, chemistry, biochemistry, biology, microbiology, and human physiology. Dietitians must become experts in the chemistry and functions of food (proteins, carbohydrates, and fats).
Through the Indigenous Lens (Suzanne Wilkerson and Charles Molnar)
I work at Camosun College located in beautiful Victoria, British Columbia with campuses on the Traditional Territories of the Lekwungen and W̱SÁNEĆ peoples. The underground storage bulb of the camas flower shown below has been an important food source for many of the Indigenous peoples of Vancouver Island and throughout the western area of North America. Camas bulbs are still eaten as a traditional food source and the preparation of the camas bulbs relates to this text section about carbohydrates.

Figure 2.16 Image of a blue camas flower and an insect pollinator. The underground bulb of camas is baked in a fire pit. Heat acts like pancreatic amylase enzyme and breaks down long chains of indigestible inulin into digestible mono and di-saccharides.
Most often plants create starch as the stored form of carbohydrate. Some plants, like camas create inulin. Inulin is used as dietary fibre however, it is not readily digested by humans. If you were to bite into a raw camas bulb it would taste bitter and has a gummy texture. The method used by Indigenous peoples to make camas both digestible and tasty is to bake the bulbs slowly for a long period in an underground firepit covered with specific leaves and soil. The heat acts like our pancreatic amylase enzyme and breaks down the long chains of inulin into digestible mono and di-saccharides.
Properly baked, the camas bulbs taste like a combination of baked pear and cooked fig. It is important to note that while the blue camas is a food source, it should not be confused with the white death camas, which is particularly toxic and deadly. The flowers look different, but the bulbs look very similar.
Lipids
Lipids include a diverse group of compounds that are united by a common feature. Lipids are hydrophobic (“water-fearing”), or insoluble in water, because they are nonpolar molecules. This is because they are hydrocarbons that include only nonpolar carbon-carbon or carbon-hydrogen bonds. Lipids perform many different functions in a cell. Cells store energy for long-term use in the form of lipids called fats. Lipids also provide insulation from the environment for plants and animals. For example, they help keep aquatic birds and mammals dry because of their water-repelling nature. Lipids are also the building blocks of many hormones and are an important constituent of the plasma membrane. Lipids include fats, oils, waxes, phospholipids, and steroids.
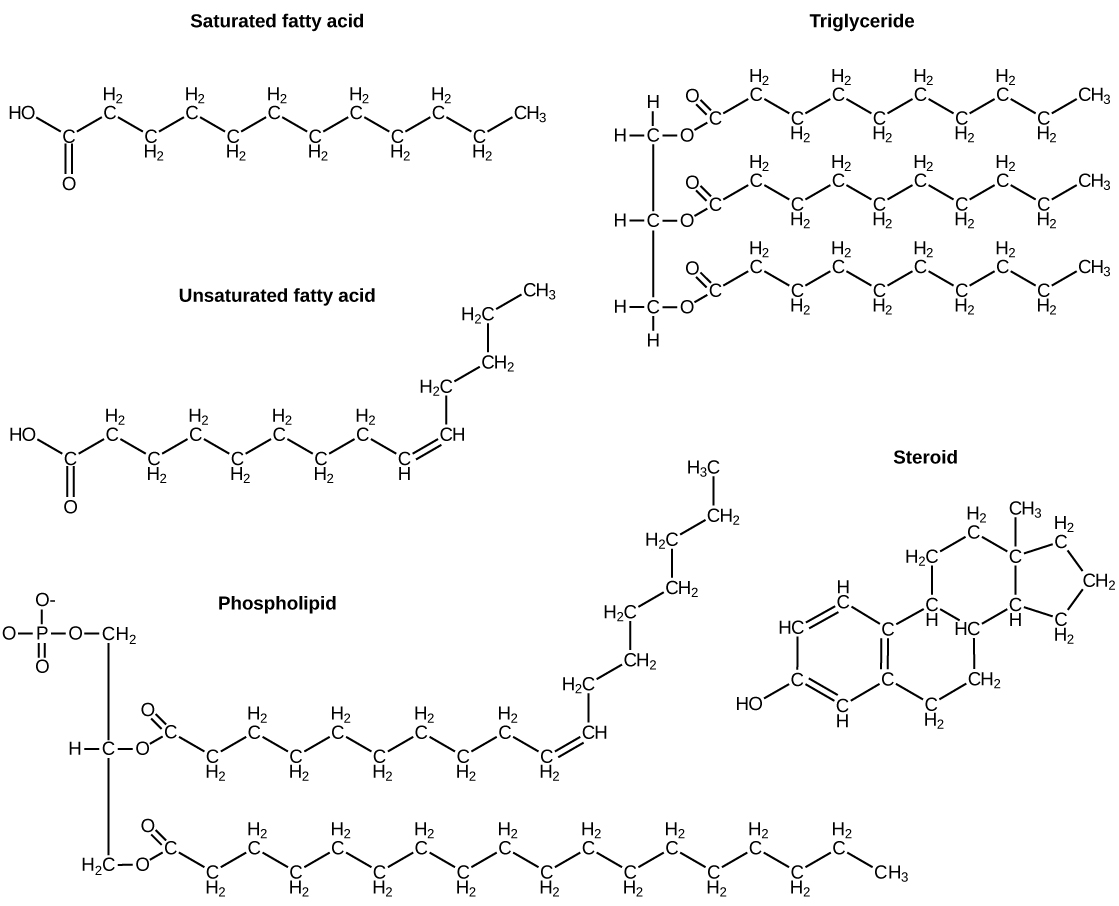
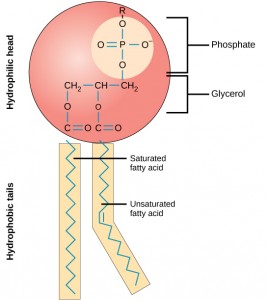
A fat molecule, such as a triglyceride, consists of two main components—glycerol and fatty acids. Glycerol is an organic compound with three carbon atoms, five hydrogen atoms, and three hydroxyl (–OH) groups. Fatty acids have a long chain of hydrocarbons to which an acidic carboxyl group is attached, hence the name “fatty acid.” The number of carbons in the fatty acid may range from 4 to 36; most common are those containing 12–18 carbons. In a fat molecule, a fatty acid is attached to each of the three oxygen atoms in the –OH groups of the glycerol molecule with a covalent bond.
During this covalent bond formation, three water molecules are released. The three fatty acids in the fat may be similar or dissimilar. These fats are also called triglycerides because they have three fatty acids. Some fatty acids have common names that specify their origin. For example, palmitic acid, a saturated fatty acid, is derived from the palm tree. Arachidic acid is derived from Arachis hypogaea, the scientific name for peanuts.
Fatty acids may be saturated or unsaturated. In a fatty acid chain, if there are only single bonds between neighboring carbons in the hydrocarbon chain, the fatty acid is saturated. Saturated fatty acids are saturated with hydrogen; in other words, the number of hydrogen atoms attached to the carbon skeleton is maximized.
When the hydrocarbon chain contains a double bond, the fatty acid is an unsaturated fatty acid.
Most unsaturated fats are liquid at room temperature and are called oils. If there is one double bond in the molecule, then it is known as a monounsaturated fat (e.g., olive oil), and if there is more than one double bond, then it is known as a polyunsaturated fat (e.g., canola oil).
Saturated fats tend to get packed tightly and are solid at room temperature. Animal fats with stearic acid and palmitic acid contained in meat, and the fat with butyric acid contained in butter, are examples of saturated fats. Mammals store fats in specialized cells called adipocytes, where globules of fat occupy most of the cell. In plants, fat or oil is stored in seeds and is used as a source of energy during embryonic development.
Unsaturated fats or oils are usually of plant origin and contain unsaturated fatty acids. The double bond causes a bend or a “kink” that prevents the fatty acids from packing tightly, keeping them liquid at room temperature. Olive oil, corn oil, canola oil, and cod liver oil are examples of unsaturated fats. Unsaturated fats help to improve blood cholesterol levels, whereas saturated fats contribute to plaque formation in the arteries, which increases the risk of a heart attack.
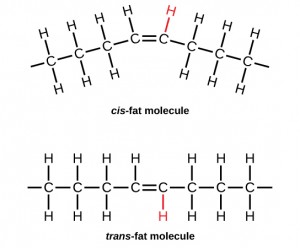
In the food industry, oils are artificially hydrogenated to make them semi-solid, leading to less spoilage and increased shelf life. Simply speaking, hydrogen gas is bubbled through oils to solidify them. During this hydrogenation process, double bonds of the cis-conformation in the hydrocarbon chain may be converted to double bonds in the trans-conformation. This forms a trans-fat from a cis-fat. The orientation of the double bonds affects the chemical properties of the fat.
Margarine, some types of peanut butter, and shortening are examples of artificially hydrogenated trans-fats. Recent studies have shown that an increase in trans-fats in the human diet may lead to an increase in levels of low-density lipoprotein (LDL), or “bad” cholesterol, which, in turn, may lead to plaque deposition in the arteries, resulting in heart disease. Many fast food restaurants have recently eliminated the use of trans-fats, and U.S. food labels are now required to list their trans-fat content.
Essential fatty acids are fatty acids that are required but not synthesized by the human body. Consequently, they must be supplemented through the diet. Omega-3 fatty acids fall into this category and are one of only two known essential fatty acids for humans (the other being omega-6 fatty acids). They are a type of polyunsaturated fat and are called omega-3 fatty acids because the third carbon from the end of the fatty acid participates in a double bond.
Salmon, trout, and tuna are good sources of omega-3 fatty acids. Omega-3 fatty acids are important in brain function and normal growth and development. They may also prevent heart disease and reduce the risk of cancer.
Like carbohydrates, fats have received a lot of bad publicity. It is true that eating an excess of fried foods and other “fatty” foods leads to weight gain. However, fats do have important functions. Fats serve as long-term energy storage. They also provide insulation for the body. Therefore, “healthy” unsaturated fats in moderate amounts should be consumed on a regular basis.
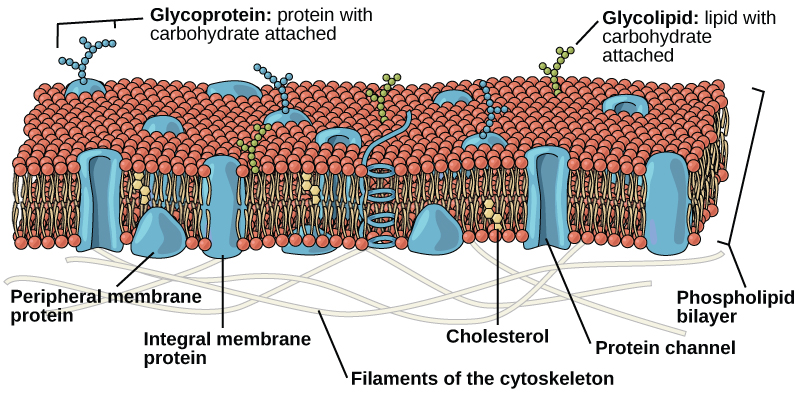
Phospholipids are the major constituent of the plasma membrane. Like fats, they are composed of fatty acid chains attached to a glycerol or similar backbone. Instead of three fatty acids attached, however, there are two fatty acids and the third carbon of the glycerol backbone is bound to a phosphate group. The phosphate group is modified by the addition of an alcohol.
A phospholipid has both hydrophobic and hydrophilic regions. The fatty acid chains are hydrophobic and exclude themselves from water, whereas the phosphate is hydrophilic and interacts with water.
Cells are surrounded by a membrane, which has a bilayer of phospholipids. The fatty acids of phospholipids face inside, away from water, whereas the phosphate group can face either the outside environment or the inside of the cell, which are both aqueous.
Through the Indigenous Lens
For the First peoples of the Pacific Northwest the fat rich fish ooligan, with 20% fat by body weight, was a crucial part of the diet of several First Nations. Why? Because fat is the most calorie dense food and having a storable, high calorie compact energy source would be important to survival. The nature of its fat also made it an important trade good. Like salmon, ooligan returns to its birth stream after years at sea. Its arrival in the early spring made it the first fresh food of the year. In the Tsimshianic languages the arrival of the ooligan … was traditionally announced with the cry, ‘Hlaa aat’ixshi halimootxw!’ … meaning ‘Our Saviour has just arrived!’

Figure 2.20 Image of cooked ooligan. With 20% fat by body weight, this fat rich fish is a crucial part of the First Nations diet.
As you learned above all fats are hydrophobic (water hating). To isolate the fat, the fish is boiled and the floating fat skimmed off. Ooligan fat composition is 30% saturated fat (like butter) and 55% monounsaturated fat (like plant oils). Importantly it is a solid grease at room temperature. Because it is low in polyunsaturated fats (which oxidize and spoil quickly) it can be stored for later use and used as a trade item. Its composition is said to make it as healthy as olive oil, or better as it has omega 3 fatty acids that reduce risk for diabetes and stroke. It also is rich in three fat soluble vitamins A, E and K.
Steroids and Waxes
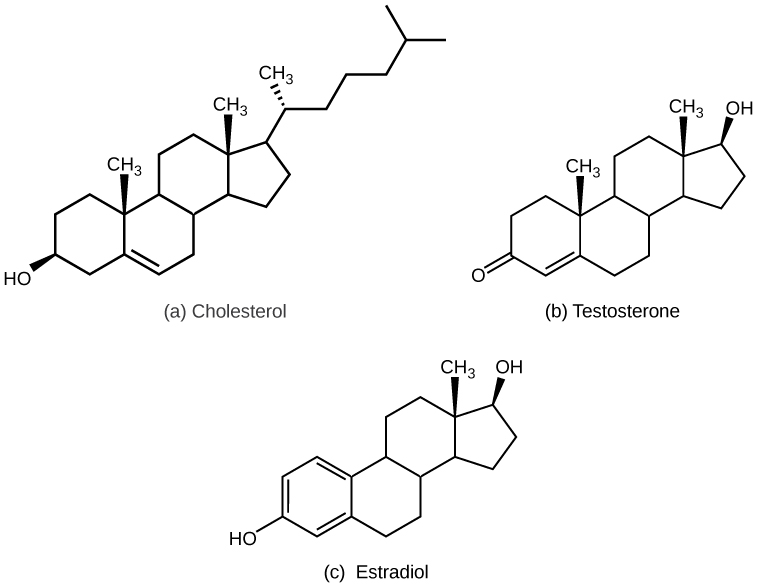
Unlike the phospholipids and fats discussed earlier, steroids have a ring structure. Although they do not resemble other lipids, they are grouped with them because they are also hydrophobic. All steroids have four, linked carbon rings and several of them, like cholesterol, have a short tail.
Cholesterol is a steroid. Cholesterol is mainly synthesized in the liver and is the precursor of many steroid hormones, such as testosterone and estradiol. It is also the precursor of vitamins E and K. Cholesterol is the precursor of bile salts, which help in the breakdown of fats and their subsequent absorption by cells. Although cholesterol is often spoken of in negative terms, it is necessary for the proper functioning of the body. It is a key component of the plasma membranes of animal cells.
Waxes are made up of a hydrocarbon chain with an alcohol (–OH) group and a fatty acid. Examples of animal waxes include beeswax and lanolin. Plants also have waxes, such as the coating on their leaves, that helps prevent them from drying out.
Concept in Action

For an additional perspective on lipids, explore “Biomolecules: The Lipids” through this interactive animation.
Proteins
Proteins are one of the most abundant organic molecules in living systems and have the most diverse range of functions of all macromolecules. Proteins may be structural, regulatory, contractile, or protective; they may serve in transport, storage, or membranes; or they may be toxins or enzymes. Each cell in a living system may contain thousands of different proteins, each with a unique function. Their structures, like their functions, vary greatly. They are all, however, polymers of amino acids, arranged in a linear sequence.
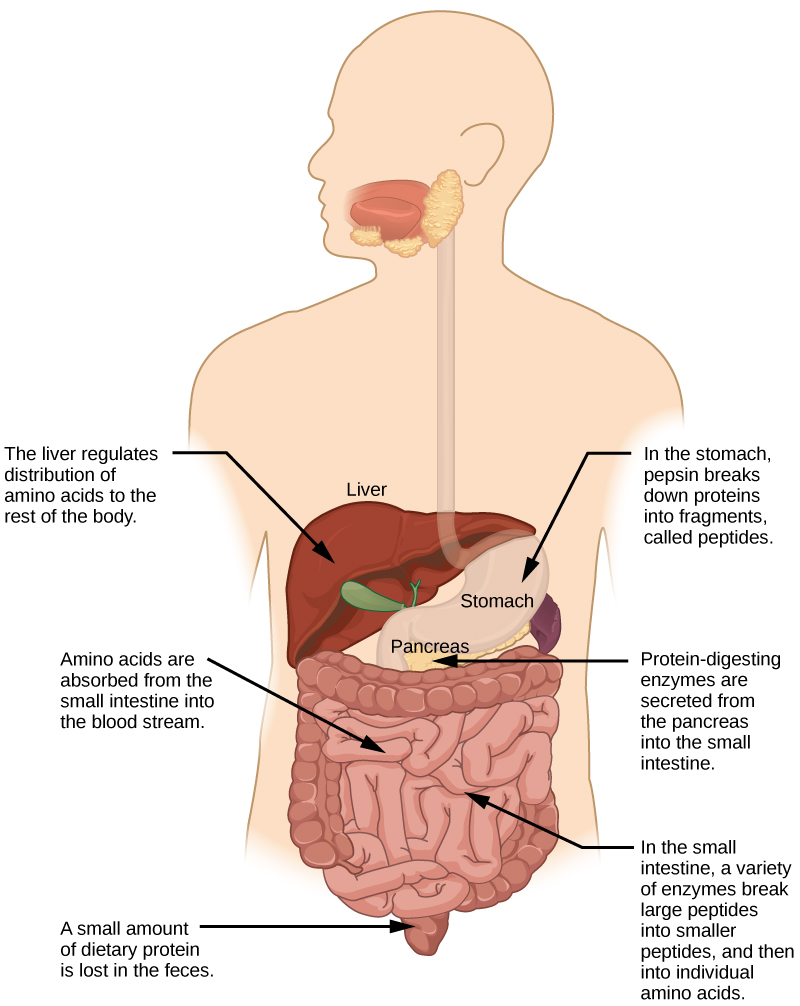
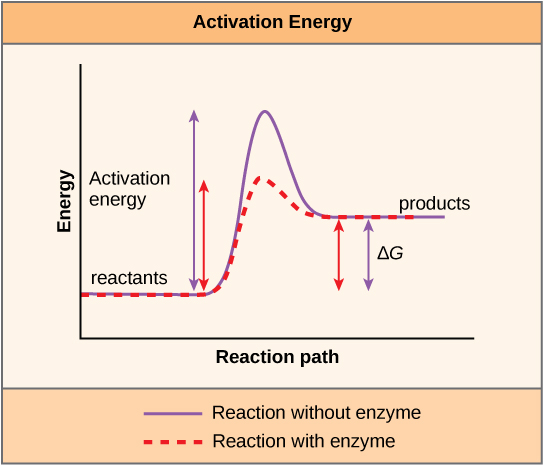
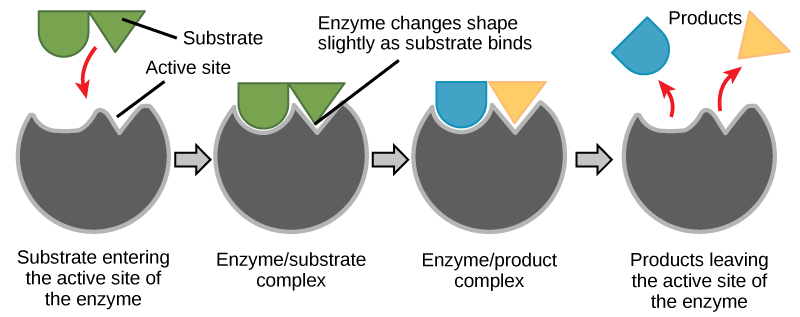
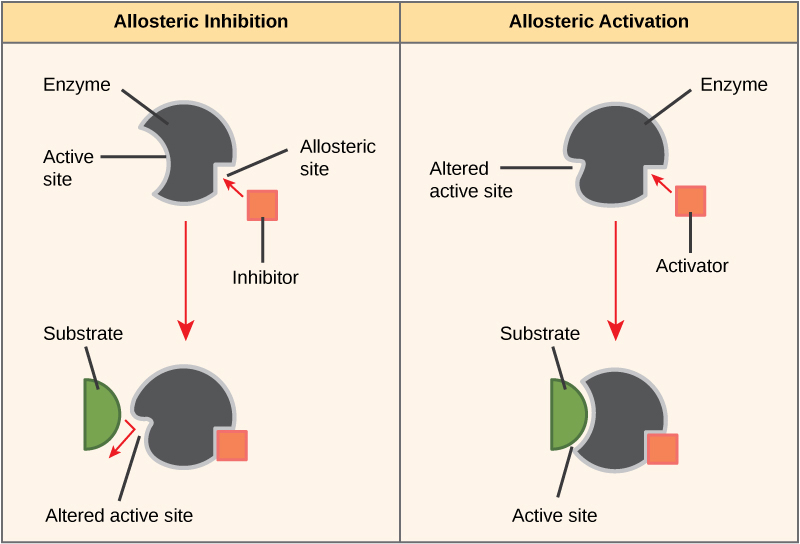
The functions of proteins are very diverse because there are 20 different chemically distinct amino acids that form long chains, and the amino acids can be in any order. For example, proteins can function as enzymes or hormones. Enzymes, which are produced by living cells, are catalysts in biochemical reactions (like digestion) and are usually proteins. Each enzyme is specific for the substrate (a reactant that binds to an enzyme) upon which it acts. Enzymes can function to break molecular bonds, to rearrange bonds, or to form new bonds. An example of an enzyme is salivary amylase, which breaks down amylose, a component of starch.
Hormones are chemical signaling molecules, usually proteins or steroids, secreted by an endocrine gland or group of endocrine cells that act to control or regulate specific physiological processes, including growth, development, metabolism, and reproduction. For example, insulin is a protein hormone that maintains blood glucose levels.
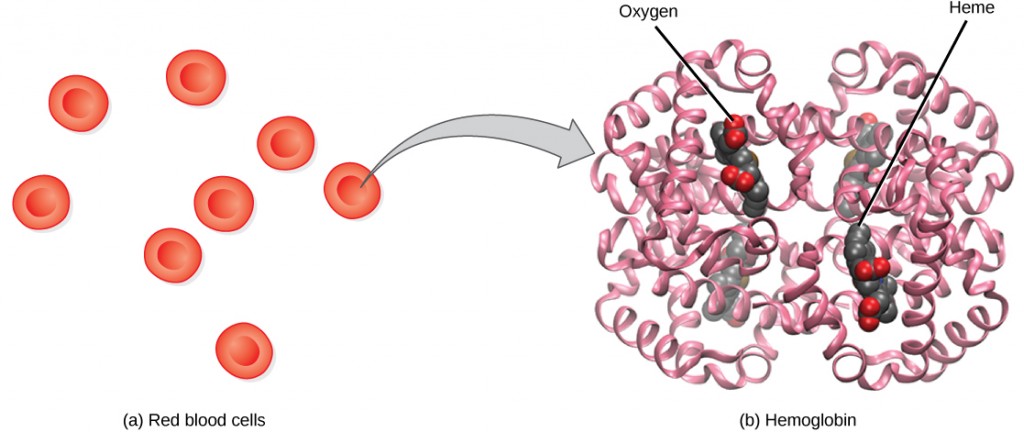
Proteins have different shapes and molecular weights; some proteins are globular in shape whereas others are fibrous in nature. For example, hemoglobin is a globular protein, but collagen, found in our skin, is a fibrous protein. Protein shape is critical to its function. Changes in temperature, pH, and exposure to chemicals may lead to permanent changes in the shape of the protein, leading to a loss of function or denaturation (to be discussed in more detail later). All proteins are made up of different arrangements of the same 20 kinds of amino acids.
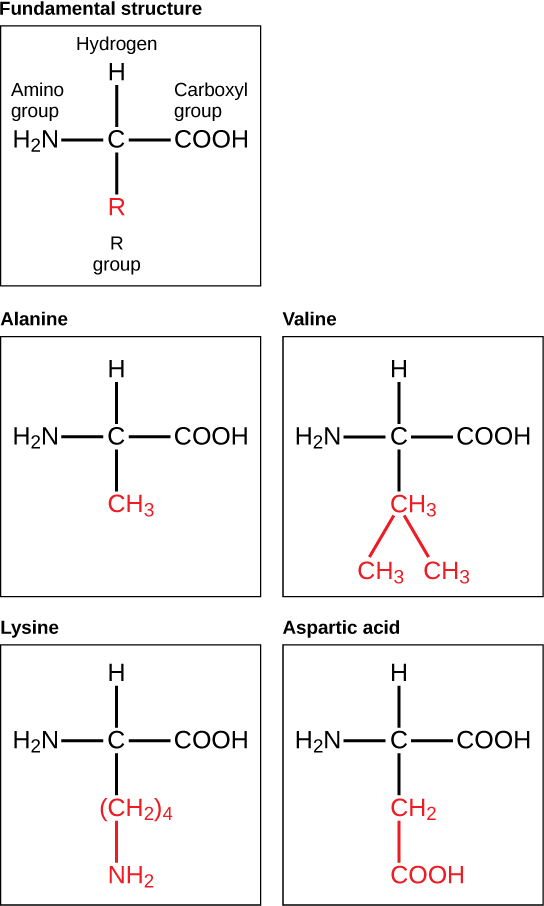
Amino acids are the monomers that make up proteins. Each amino acid has the same fundamental structure, which consists of a central carbon atom bonded to an amino group (–NH2), a carboxyl group (–COOH), and a hydrogen atom. Every amino acid also has another variable atom or group of atoms bonded to the central carbon atom known as the R group. The R group is the only difference in structure between the 20 amino acids; otherwise, the amino acids are identical.
The chemical nature of the R group determines the chemical nature of the amino acid within its protein (that is, whether it is acidic, basic, polar, or nonpolar).
The sequence and number of amino acids ultimately determine a protein’s shape, size, and function. Each amino acid is attached to another amino acid by a covalent bond, known as a peptide bond, which is formed by a dehydration reaction. The carboxyl group of one amino acid and the amino group of a second amino acid combine, releasing a water molecule. The resulting bond is the peptide bond.
The products formed by such a linkage are called polypeptides. While the terms polypeptide and protein are sometimes used interchangeably, a polypeptide is technically a polymer of amino acids, whereas the term protein is used for a polypeptide or polypeptides that have combined together, have a distinct shape, and have a unique function.
Evolution in Action
The Evolutionary Significance of Cytochrome cCytochrome c is an important component of the molecular machinery that harvests energy from glucose. Because this protein’s role in producing cellular energy is crucial, it has changed very little over millions of years. Protein sequencing has shown that there is a considerable amount of sequence similarity among cytochrome c molecules of different species; evolutionary relationships can be assessed by measuring the similarities or differences among various species’ protein sequences.
For example, scientists have determined that human cytochrome c contains 104 amino acids. For each cytochrome c molecule that has been sequenced to date from different organisms, 37 of these amino acids appear in the same position in each cytochrome c. This indicates that all of these organisms are descended from a common ancestor. On comparing the human and chimpanzee protein sequences, no sequence difference was found. When human and rhesus monkey sequences were compared, a single difference was found in one amino acid. In contrast, human-to-yeast comparisons show a difference in 44 amino acids, suggesting that humans and chimpanzees have a more recent common ancestor than humans and the rhesus monkey, or humans and yeast.
Protein Structure
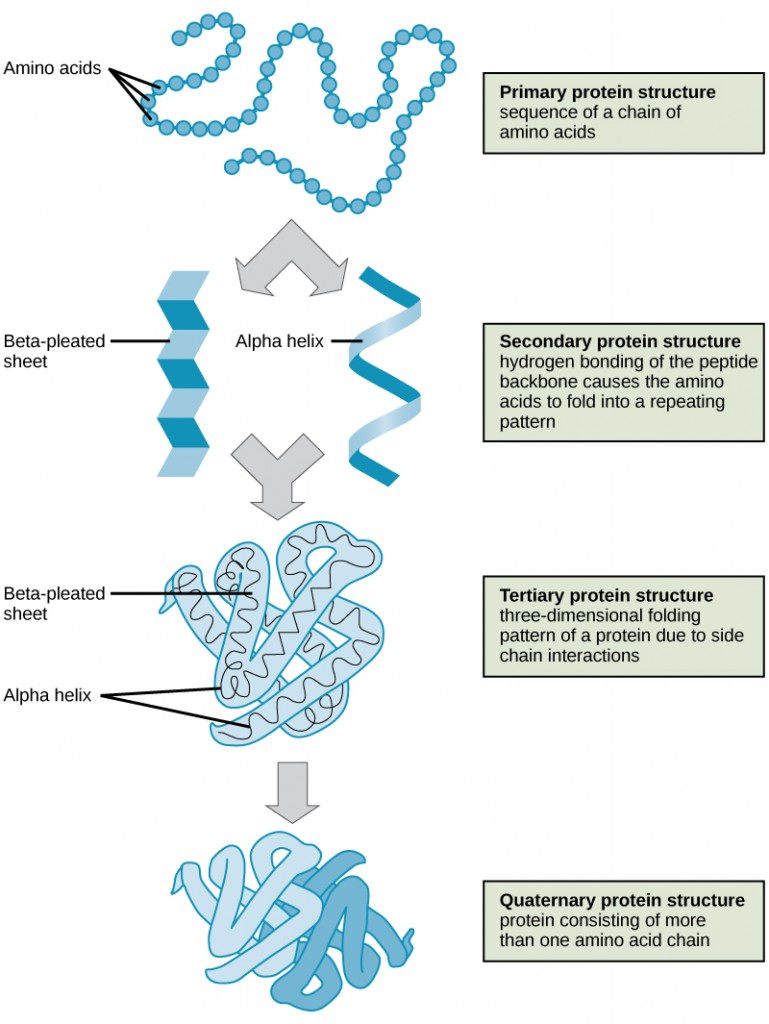
As discussed earlier, the shape of a protein is critical to its function. To understand how the protein gets its final shape or conformation, we need to understand the four levels of protein structure: primary, secondary, tertiary, and quaternary.
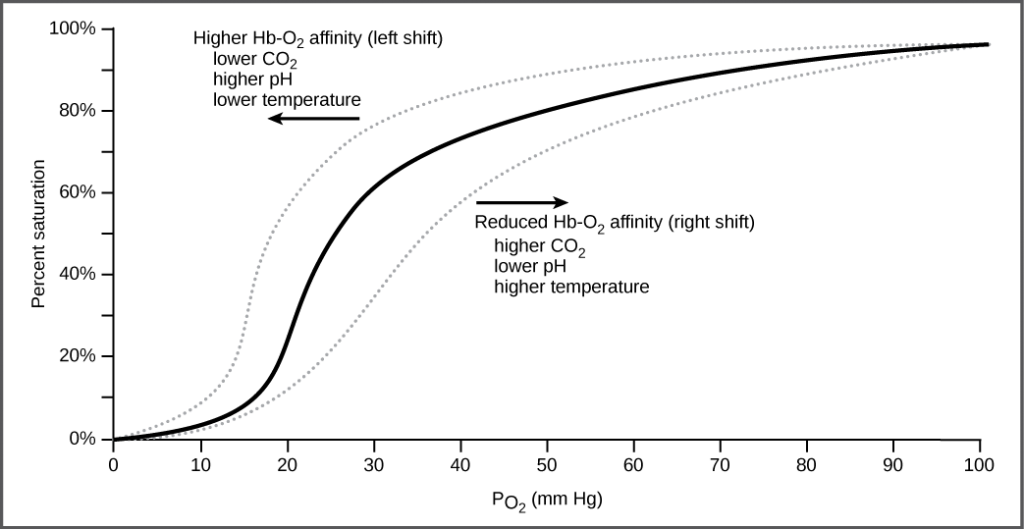
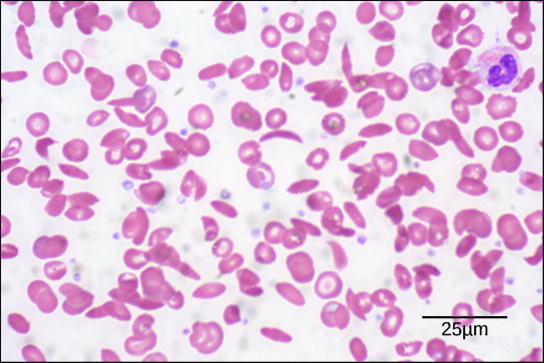
The unique sequence and number of amino acids in a polypeptide chain is its primary structure. The unique sequence for every protein is ultimately determined by the gene that encodes the protein. Any change in the gene sequence may lead to a different amino acid being added to the polypeptide chain, causing a change in protein structure and function. In sickle cell anemia, the hemoglobin β chain has a single amino acid substitution, causing a change in both the structure and function of the protein. What is most remarkable to consider is that a hemoglobin molecule is made up of two alpha chains and two beta chains that each consist of about 150 amino acids. The molecule, therefore, has about 600 amino acids. The structural difference between a normal hemoglobin molecule and a sickle cell molecule—that dramatically decreases life expectancy in the affected individuals—is a single amino acid of the 600.
Because of this change of one amino acid in the chain, the normally biconcave, or disc-shaped, red blood cells assume a crescent or “sickle” shape, which clogs arteries. This can lead to a myriad of serious health problems, such as breathlessness, dizziness, headaches, and abdominal pain for those who have this disease.
Folding patterns resulting from interactions between the non-R group portions of amino acids give rise to the secondary structure of the protein. The most common are the alpha (α)-helix and beta (β)-pleated sheet structures. Both structures are held in shape by hydrogen bonds. In the alpha helix, the bonds form between every fourth amino acid and cause a twist in the amino acid chain.
In the β-pleated sheet, the “pleats” are formed by hydrogen bonding between atoms on the backbone of the polypeptide chain. The R groups are attached to the carbons, and extend above and below the folds of the pleat. The pleated segments align parallel to each other, and hydrogen bonds form between the same pairs of atoms on each of the aligned amino acids. The α-helix and β-pleated sheet structures are found in many globular and fibrous proteins.
The unique three-dimensional structure of a polypeptide is known as its tertiary structure. This structure is caused by chemical interactions between various amino acids and regions of the polypeptide. Primarily, the interactions among R groups create the complex three-dimensional tertiary structure of a protein. There may be ionic bonds formed between R groups on different amino acids, or hydrogen bonding beyond that involved in the secondary structure. When protein folding takes place, the hydrophobic R groups of nonpolar amino acids lay in the interior of the protein, whereas the hydrophilic R groups lay on the outside. The former types of interactions are also known as hydrophobic interactions.
In nature, some proteins are formed from several polypeptides, also known as subunits, and the interaction of these subunits forms the quaternary structure. Weak interactions between the subunits help to stabilize the overall structure. For example, hemoglobin is a combination of four polypeptide subunits.
Each protein has its own unique sequence and shape held together by chemical interactions. If the protein is subject to changes in temperature, pH, or exposure to chemicals, the protein structure may change, losing its shape in what is known as denaturation as discussed earlier. Denaturation is often reversible because the primary structure is preserved if the denaturing agent is removed, allowing the protein to resume its function. Sometimes denaturation is irreversible, leading to a loss of function. One example of protein denaturation can be seen when an egg is fried or boiled. The albumin protein in the liquid egg white is denatured when placed in a hot pan, changing from a clear substance to an opaque white substance. Not all proteins are denatured at high temperatures; for instance, bacteria that survive in hot springs have proteins that are adapted to function at those temperatures.
Concept in Action

For an additional perspective on proteins, explore “Biomolecules: The Proteins” through this interactive animation.
Nucleic Acids
Nucleic acids are key macromolecules in the continuity of life. They carry the genetic blueprint of a cell and carry instructions for the functioning of the cell.
The two main types of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). DNA is the genetic material found in all living organisms, ranging from single-celled bacteria to multicellular mammals.
The other type of nucleic acid, RNA, is mostly involved in protein synthesis. The DNA molecules never leave the nucleus, but instead use an RNA intermediary to communicate with the rest of the cell. Other types of RNA are also involved in protein synthesis and its regulation.
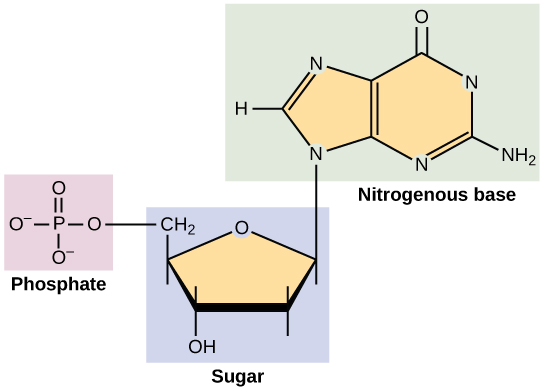
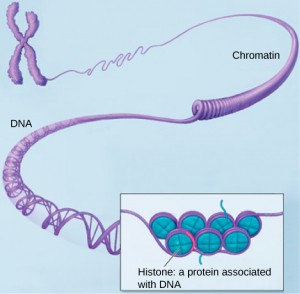
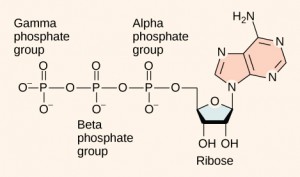
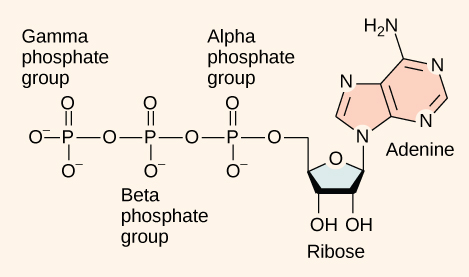
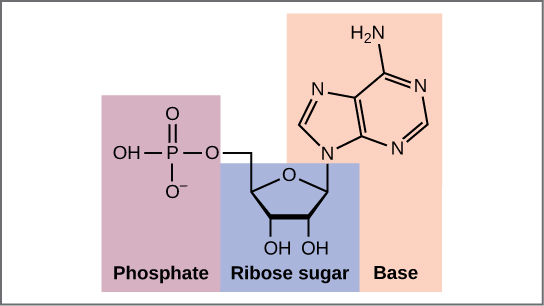
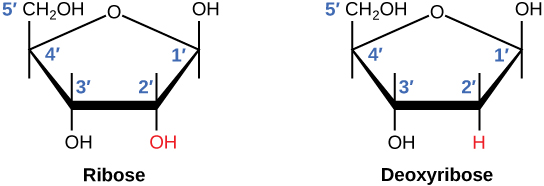
DNA and RNA are made up of monomers known as nucleotides. The nucleotides combine with each other to form a polynucleotide, DNA or RNA. Each nucleotide is made up of three components: a nitrogenous base, a pentose (five-carbon) sugar, and a phosphate group . Each nitrogenous base in a nucleotide is attached to a sugar molecule, which is attached to a phosphate group.
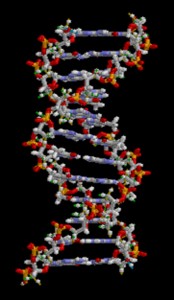
DNA Double-Helical Structure
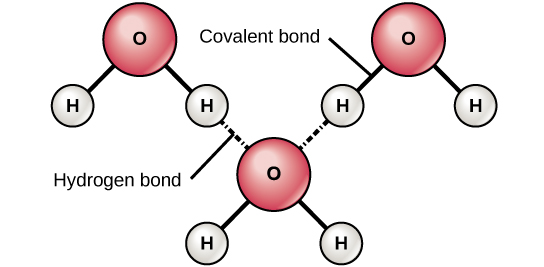
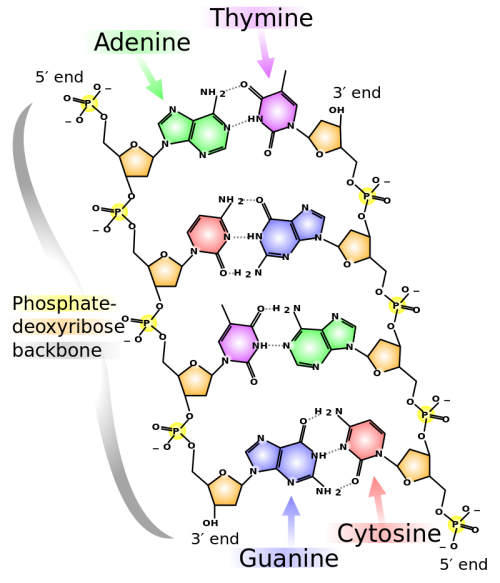
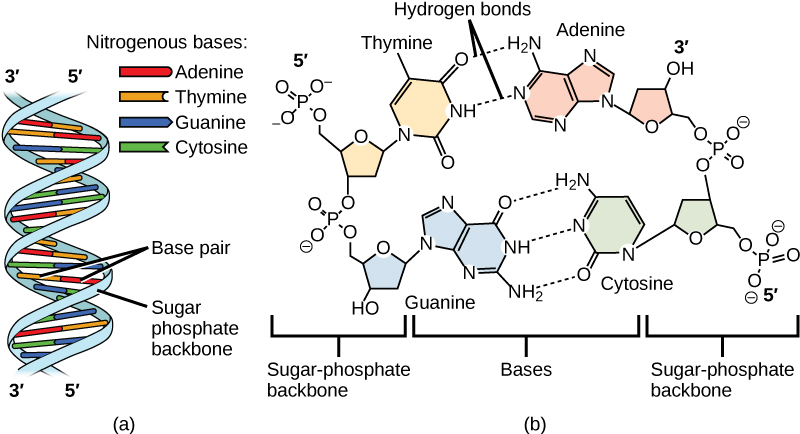
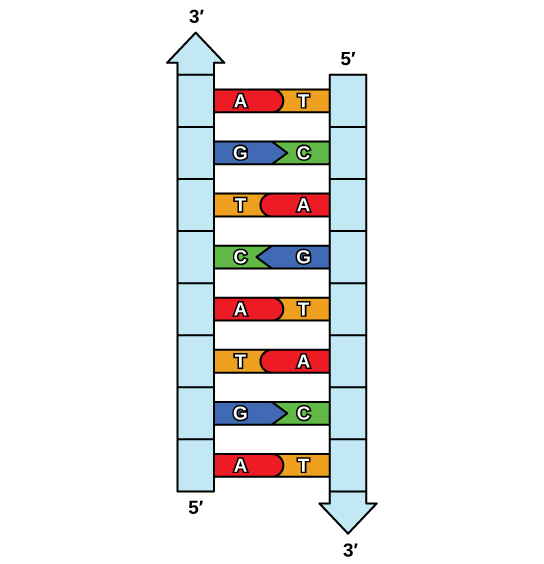
DNA has a double-helical structure. It is composed of two strands, or polymers, of nucleotides. The strands are formed with bonds between phosphate and sugar groups of adjacent nucleotides. The strands are bonded to each other at their bases with hydrogen bonds, and the strands coil about each other along their length, hence the “double helix” description, which means a double spiral.
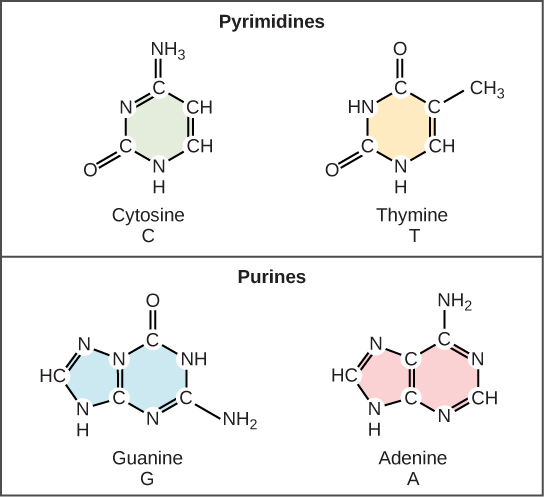
The alternating sugar and phosphate groups lie on the outside of each strand, forming the backbone of the DNA. The nitrogenous bases are stacked in the interior, like the steps of a staircase, and these bases pair; the pairs are bound to each other by hydrogen bonds. The bases pair in such a way that the distance between the backbones of the two strands is the same all along the molecule. The rule is that nucleotide A pairs with nucleotide T, and G with C, see section 9.1 for more details.
Section Summary
Living things are carbon-based because carbon plays such a prominent role in the chemistry of living things. The four covalent bonding positions of the carbon atom can give rise to a wide diversity of compounds with many functions, accounting for the importance of carbon in living things. Carbohydrates are a group of macromolecules that are a vital energy source for the cell, provide structural support to many organisms, and can be found on the surface of the cell as receptors or for cell recognition. Carbohydrates are classified as monosaccharides, disaccharides, and polysaccharides, depending on the number of monomers in the molecule.
Lipids are a class of macromolecules that are nonpolar and hydrophobic in nature. Major types include fats and oils, waxes, phospholipids, and steroids. Fats and oils are a stored form of energy and can include triglycerides. Fats and oils are usually made up of fatty acids and glycerol.
Proteins are a class of macromolecules that can perform a diverse range of functions for the cell. They help in metabolism by providing structural support and by acting as enzymes, carriers or as hormones. The building blocks of proteins are amino acids. Proteins are organized at four levels: primary, secondary, tertiary, and quaternary. Protein shape and function are intricately linked; any change in shape caused by changes in temperature, pH, or chemical exposure may lead to protein denaturation and a loss of function.
Nucleic acids are molecules made up of repeating units of nucleotides that direct cellular activities such as cell division and protein synthesis. Each nucleotide is made up of a pentose sugar, a nitrogenous base, and a phosphate group. There are two types of nucleic acids: DNA and RNA.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://opentextbc.ca/biology/?p=4368#h5p-15
amino acid: a monomer of a protein
carbohydrate: a biological macromolecule in which the ratio of carbon to hydrogen to oxygen is 1:2:1; carbohydrates serve as energy sources and structural support in cells
cellulose: a polysaccharide that makes up the cell walls of plants and provides structural support to the cell
chitin: a type of carbohydrate that forms the outer skeleton of arthropods, such as insects and crustaceans, and the cell walls of fungi
denaturation: the loss of shape in a protein as a result of changes in temperature, pH, or exposure to chemicals
deoxyribonucleic acid (DNA): a double-stranded polymer of nucleotides that carries the hereditary information of the cell
disaccharide: two sugar monomers that are linked together by a peptide bond
enzyme: a catalyst in a biochemical reaction that is usually a complex or conjugated protein
fat: a lipid molecule composed of three fatty acids and a glycerol (triglyceride) that typically exists in a solid form at room temperature
glycogen: a storage carbohydrate in animals
hormone: a chemical signaling molecule, usually a protein or steroid, secreted by an endocrine gland or group of endocrine cells; acts to control or regulate specific physiological processes
lipids: a class of macromolecules that are nonpolar and insoluble in water
macromolecule: a large molecule, often formed by polymerization of smaller monomers
monosaccharide: a single unit or monomer of carbohydrates
nucleic acid: a biological macromolecule that carries the genetic information of a cell and carries instructions for the functioning of the cell
nucleotide: a monomer of nucleic acids; contains a pentose sugar, a phosphate group, and a nitrogenous base
oil: an unsaturated fat that is a liquid at room temperature
phospholipid: a major constituent of the membranes of cells; composed of two fatty acids and a phosphate group attached to the glycerol backbone
polypeptide: a long chain of amino acids linked by peptide bonds
polysaccharide: a long chain of monosaccharides; may be branched or unbranched
protein: a biological macromolecule composed of one or more chains of amino acids
ribonucleic acid (RNA): a single-stranded polymer of nucleotides that is involved in protein synthesis
saturated fatty acid: a long-chain hydrocarbon with single covalent bonds in the carbon chain; the number of hydrogen atoms attached to the carbon skeleton is maximized
starch: a storage carbohydrate in plants
steroid: a type of lipid composed of four fused hydrocarbon rings
trans-fat: a form of unsaturated fat with the hydrogen atoms neighboring the double bond across from each other rather than on the same side of the double bond
triglyceride: a fat molecule; consists of three fatty acids linked to a glycerol molecule
unsaturated fatty acid: a long-chain hydrocarbon that has one or more than one double bonds in the hydrocarbon chain
Media Attribution













































































![A diagram showing a human chromosome with bands revealed with a Giemsa stain. The bands are labeled with Xp and a number on the short arm and Xq and a number on the long arm. Certain genes are found within some of the bands. These genes are labeled on the right: Fanconi anemia B, Wiskott-Aldrich syndrome, Pelizaeus-Merzbacher disease, Fragile X syndrome, and G6PD deficiency[0].](https://opentextbc.ca/biology/wp-content/uploads/sites/96/2015/10/Figure_10_03_01-445x1024-1.jpg)